Notes from the Lab: Hunting for Bacteria in Our Lab
On any given day ARAC scientists are working on multiple research projects, so we wanted to spotlight what's going on in the lab! We’ve spent much of the summer preparing for a big project that we’re kicking off this month. In short, over the next three years, we are measuring changes in three key bacteria (E. coli, Salmonella and Campylobacter) on retail chicken meat sold in California and comparing those to the bacteria infecting people.
The first step in this project is collecting bacteria from retail chicken. There are lots of bacteria on retail chicken, but what we’re looking specifically at the kinds that most commonly cause antibiotic-resistant infections in people. Here’s a look at what our researchers will be doing day in and day out as they process thousands of pieces of raw chicken over the next three years. The steps outlined below take roughly three weeks to run through all four steps. This process will be repeated over and over again until the project is completed.
Step 1: Create and test protocols for each bacterium because different bacteria thrive under different conditions. For example: Campylobacter likes to grow in a broth supplemented with a little blood while Salmonella and E. coli prefer broths supplemented with different sugars and proteins. Protocols are also created for extracting DNA, confirming the identity of the bacteria, testing for antibiotic resistance, and even shipping and receiving bacteria samples from our collaborators in California. (And everything is done in a way that ensures the safety of our team.)
Step 2: Our lab receives more than a hundred packages of chicken per month, which we then enrich in order to find our target bacteria.
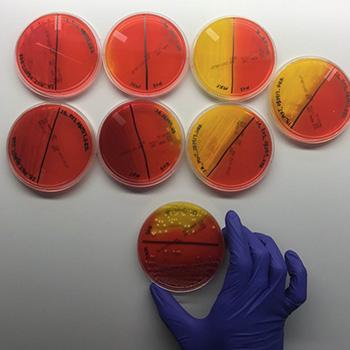
Step 3: Next, we isolate our target bacteria from the various broths by spreading them multiple times on petri dishes to separate them from other bacteria that may have grown in the broth. The petri dishes are filled with a jello-like substance called agar. Each agar is specifically formulated to encourage the growth of our target bacteria and some even make the bacteria change colors to help us find them. For example, on one of the agars that we use, E. coli colonies turn bright pink so we can easily differentiate them from other closely related bacteria.
After we’ve done all this work, we want to create a databank of each isolate, so we instantly freeze some of our purified bacteria in a liquid nitrogen bath to preserve them for future analyses (kind of like they did to Austin Powers -- yeah baby!).
Step 4: In order to make sure that we’re working with the right bacteria, we use something called “real-time PCR” to confirm that the DNA belongs to the species we think it does (i.e., Salmonella, Campylobacter, or E. coli) . So yes, even with all of these previous steps, we have to confirm our findings at the molecular level.
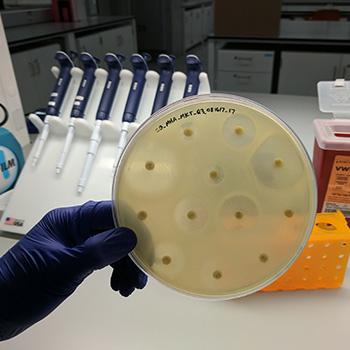
Step 5: Lastly, we conduct antibiotic susceptibility testing. Our researchers take the purified isolates and will grow a big plate of it to measure how resistant that particular bacteria is. For example in the picture below, there are 12 antibiotics (the little round disks) tested on a plate of E. coli. The antibiotics that are effective against the bacteria keep it from growing near the disks. Antibiotics that are not effective won’t stop the growth, so the bacteria will grow right up to the disks. This is seen in 3 of the disks in the picture. This bacterium is resistant to three antibiotics - which is exactly what we’re looking for!